IHC is Back!
Note to self: IHC is back in DG production! What’s IHC, you ask?
IHC is my Islandora Health Check drush command/module. Since this is a note to myself, I’m not going to get into a lot of detail here, just posting some reminders so I can remember how it works and how it is used.
What Is It?
IHC is a module that provides drush commands that can be used to collect (drush ihcC), analyze (drush ihcA), and subsequently report (drush ihcR) key object info from DG’s FEDORA repository.
At its core, the module consists of three parts:
ihcCollect(aliasihcC) collects information by walking through a user-specified range of PIDs and making calls to PHP functions, peering into and harvesting information from those objects’ XML datastreams.ihcAnalyze(aliasihcA), the analysis command, can be used to analyze the data collected byihcC. It looks at values for specific fields and for trends in the data, then applies special color or text formatting codes to highlight that data or trend.ihcReport(aliasihcR), the reporting command, is then used to output the collected and analyzed data into a readable form. Before ISLE the reports were written directly into Excel format. Since the Excel libraries I used years ago are no longer available, due to security concerns, I made theihcRcommand output data into a simple.csvformat.
History
IHC is a module (on DGDocker1 it lives in /var/www/html/sites/all/modules/islandora/ihc) that I wrote several years ago, but one that was broken with the introduction of ISLE in about 2018. In February 2022 I managed to re-work some of the code to bring it back-to-life in DG’s ISLE environment.
ihcQuick - A “Quick” Report
Running a series of ihcC, ihcA, then ihcR commands is a hassle. So, I created the drush ihcQuick (alias ihcQ) command, and it is how I recommend generating an IHC report now. A typical run of ihcQ from DG’s production node, DGDocker1, looks like this (truncated here for readability):
[islandora@dgdocker1 ~]$ docker exec -it isle-apache-dg bash
root@7f4311ed759c:/# cd /var/www/html/sites/default/
root@7f4311ed759c:/var/www/html/sites/default# drush -u 1 ihcQ grinnell:1-50000 --verbose
Executing: mysql --defaults-extra-file=/tmp/drush_FURANC --database=digital_grinnell --host=mysql --silent < /tmp/drush_lhQcPD
Executing: mysql --defaults-extra-file=/tmp/drush_Va3GiE --database=digital_grinnell --host=mysql --silent < /tmp/drush_Fode6D
icu_drush_prep will consider only objects modified after 2000-01-01T00:00:00Z. [status]
Starting operation for PID 'grinnell:1' and --repeat='49999' at 11:29:05. [status]
Fetching all valid object PIDs in the specified range. [status]
Completed fetch of 22947 valid object PIDs. [status]
Progress: ihcCollect
icu_Connect: Connection to Fedora repository as 'System Admin' is complete. [status]
pid is: grinnell:45 [status]
pid is: grinnell:47 [status]
pid is: grinnell:49 [status]
pid is: grinnell:50 [status]
...
As you can see above, it begins by shelling in to DG’s isle-apache-dg container, moving into the /var/www/html/sites/default directory, and executing the drush command of the form: drush -u 1 ihcQ grinnell:1-50000 --verbose.
This command will run a “quick” report for PIDs in the grinnell namespace with numeric IDs in the range 1 to 50000, and it will generate --verbose output.
In order to capture a complete picture or an ihcQ run I’m going to omit the --verbose option and vastly shrink the number of objects to report on, like so:
root@7f4311ed759c:/var/www/html/sites/default# drush -u 1 ihcQ grinnell:1-50
icu_drush_prep will consider only objects modified after 2000-01-01T00:00:00Z. [status]
Starting operation for PID 'grinnell:1' and --repeat='49' at 11:35:39. [status]
Fetching all valid object PIDs in the specified range. [status]
Completed fetch of 4 valid object PIDs. [status]
Progress: ihcCollect
icu_Connect: Connection to Fedora repository as 'System Admin' is complete. [status]
[=================================================================================] 100%
Completed ihcCollect at 11:35:50. Use 'drush ihcA' to analyze and 'drush ihcR' [status]
to report it.
icu_drush_prep will consider only objects modified after 2000-01-01T00:00:00Z. [status]
Starting operation for PID 'grinnell:1' and --repeat='49' at 11:35:50. [status]
Fetching all valid object PIDs in the specified range. [status]
Completed fetch of 4 valid object PIDs. [status]
Progress: ihcAnalyze
[=================================================================================] 100%
Completed ihcAnalyze at 11:36:00. Use 'drush ihcR' to report it. [ok]
icu_drush_prep will consider only objects modified after 2000-01-01T00:00:00Z. [status]
Starting operation for PID 'grinnell:1' and --repeat='49' at 11:36:00. [status]
Fetching all valid object PIDs in the specified range. [status]
Completed fetch of 4 valid object PIDs. [status]
.csv file path is: 'public://ihcQ_grinnell_ih.csv'. [status]
Progress: ihcReport
[=================================================================================] 100%
As you can see above, the ihcQ command did the following:
- Collected data by invoking an
ihcC, orihcCollect, operation; - Analyzed the data via
ihcA, orihcAnalyze; and - Reported the data using
ihcR, orihcReport.
Near the end of the output you can see that a new public://ihcQ_grinnell_ih.csv was generated.
Downloading IHC Reports
Since the IHC report is a .csv file it’s not much use sitting in the public:// folder on the server, so I use the Islandora Health Check command in the Management menu on DG’s home page to facilitate download of these reports. Clicking that command link in my browser brings me to https://digital.grinnell.edu/ihc where I’m presented with a list of _ih.csv files to download.
Once I’ve selected and downloaded one of these files I engage a Python utility of my own creation to interpret the color and format data that ihcAnalyze generated and produce a visually effective Excel output file.
Convert .csv to Excel with Formatting
On my Mac I’m able to run my format-csv-to-excel Python script like so:
╭─mcfatem@MAC02FK0XXQ05Q ~/Downloads
╰─$ python3 /Users/mcfatem/GitHub/format-csv-to-excel/main.py --verbose --dg ihcQ_grinnell_ih.csv
Verbose output selected.
Special Digital.Grinnell processing is selected.
ihcQ_grinnell_ih.xlsx has been created.
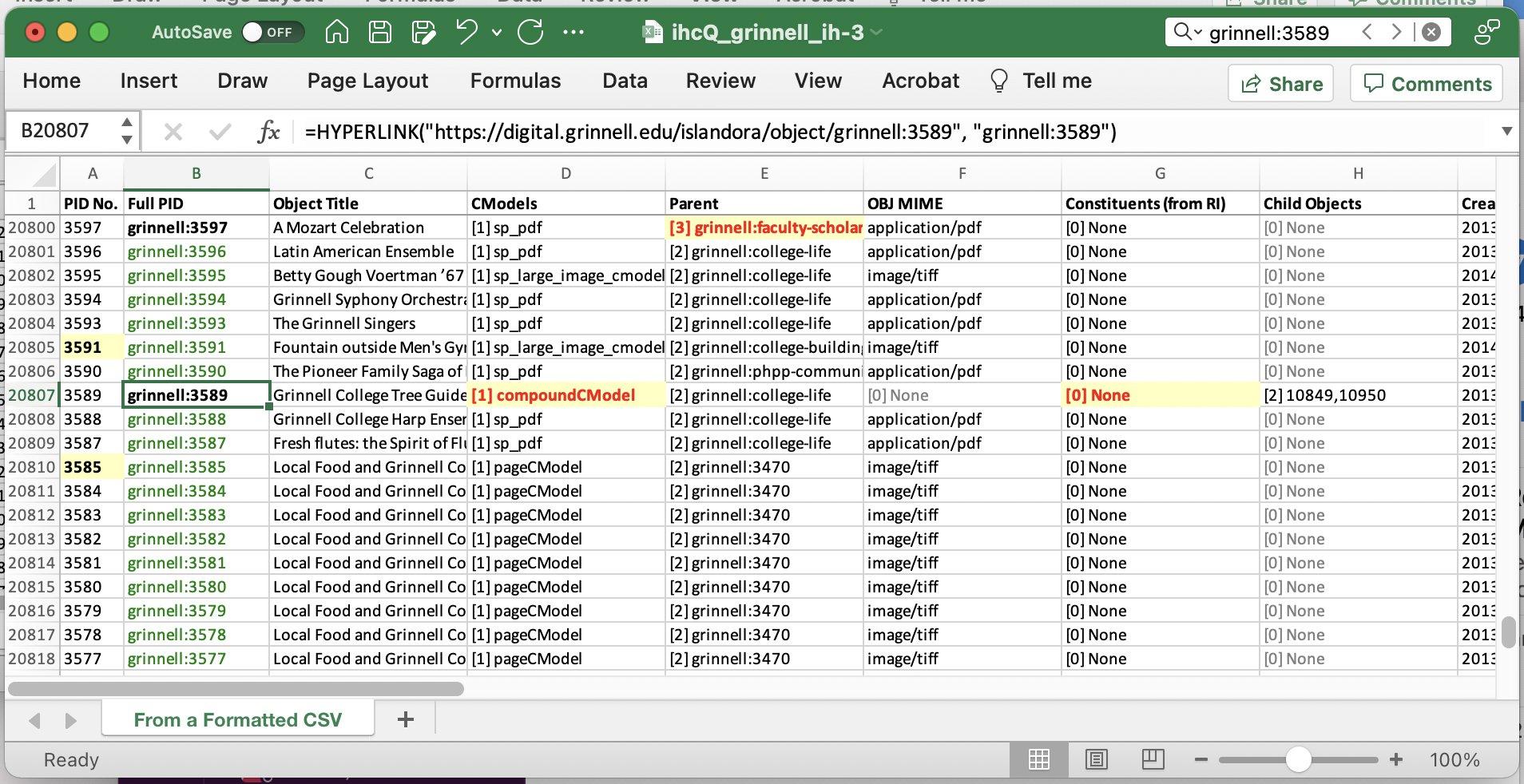
The Result
The results, in Excel, look something like the figure you see below.
And that’s a wrap. Until next time, stay safe and wash your hands! 😄